New CiPA cardiac ion channel cell lines and assays for in vitro proarrhythmia risk assessment
Case Study
Case Study title
New CiPA cardiac ion channel cell lines and assays for in vitro proarrhythmia risk assessment
Authors
Edward S.A HUMPHRIES¹, Robert W. KIRBY¹, John RIDLEY¹, Hironori OHSHIRO², and Marc ROGERS¹
- Metrion Biosciences Ltd, Riverside 3; Suite 1, Granta Park, Cambridge, CB21 6AD, United Kingdom
- Sophion Bioscience K.K., Incubation on Campus Honjo Waseda #V203 1011 Nishitomida, Honjo, Saitama 367-0035, Japan
Introduction
New cardiac safety testing guidelines are being finalised, as part of the FDA’s Comprehensive in vitro Proarrhythmia Assay (CiPA) initiative, which aim to remove the over-reliance on screening against the hERG channel by expanding the panel to include hNav1.5, hCav1.2, hKv4.3/KChiP2.2, hKir2.1 and hKv7.1/KCNE1 human cardiac ion channels. In addition, the CiPA working groups have recently identified two additional in vitro assays required for in silico models to reliably predict proarrhythmia. The first is a ‘late’ sodium current assay, as inhibition of persistent inward current can affect repolarisation and mitigate proarrhythmia (e.g. ranolazine). The second assay quantifies the degree of drug trapping in the hERG channel using the Milnes voltage protocol⁽¹⁾, which can improve the prediction of proarrhythmic risk⁽²⁾.
We have validated these additional cardiac assays on the gigaseal quality QPatch 48 automated patch clamp platform. Rather than rely on pharmacological activators such as ATX-II or veratridine to induce late openings of the hNav1.5 channel, we created a HEK cell line expressing the LQT3 ΔKPQ hNav1.5 mutant which promotes activation of persistent sodium current⁽³⁾. Late sodium current was measured using step pulse, step-ramp, ramp, and action potential waveform voltage protocols, using proprietary solutions to minimise rundown and increase whole-cell seal duration.
hNav1.5 ΔKPQ cells exhibited ranolazine-sensitive late sustained inward currents that were ~1-3% magnitude of the peak current (>4 nA) in single hole QPlate 48 recordings. A CHO cell line expressing the hERG channel and the published Milnes hERG kinetic protocol were optimised for the QPatch 48 to create a biophysically and pharmacologically validated ‘dynamic’ hERG assay. We compensated for slower activation kinetics at room temperature (compared with manual patch at 37 °C) and optimised liquid addition and voltage protocols to create a stable assay during prolonged vehicle applications. Our QPatch 48 Milnes hERG assay correctly reports the drug trapping profiles of cisapride (10%) and dofetilide (95%), as well as several other CiPA toolbox reference compounds.
We developed a hNav1.5 ΔKPQ mutant cell line and a Milnes dynamic hERG kinetic assay on a gigaseal quality screening platform that meet the additional requirements of the CiPA initiative, and can provide high quality HTS automated patch clamp data for more accurate cardiac safety assessment.
Materials and Methods
HEK293 and CHO cells were transfected using standard liposomal transfection reagents with hNav1.5 (ΔKPQ) mutant cDNA obtained commercially and verified by sequencing. All data are from single hole chips. Standard QPatch cell suspension, sealing and wholecell protocols were utilized, with minor adjustments to obtain a high proportion of gigaohm seals.
Dynamic hERG assay
CHO cells expressing the human ether-a-go-go related potassium channel (hERG, Kv11.1) were obtained from B’SYS. Cells were cultured and harvested using our optimised QPatch protocols. Standard QPatch cell suspension, sealing and whole-cell protocols were utilized, with minor adjustments to obtain a high proportion of gigaohm seals and acceptable whole-cell hERG current amplitude, and stable current kinetics.
An additional CiPA channel component required for accurately predicting proarrhythmic liability is the ‘late’ or persistent sodium current. This small current persists throughout the cardiac action potential after initial inactivation of over 99% of sodium channels⁽⁴⁾. The small amplitude of the wildtype late current is not amenable to automated patch clamp recordings, so activators, such as ATXII and veratridine, have been used induce late openings. However, large shifts in IC50 values for drugs, such as ranolazine, occur between each activator, which can also open endogenous sodium currents.
Metrion aimed to remove the need to activate the ‘late’ hNav1.5 current using non-specific pharmacological tools by creating a cell line expressing a long QT syndrome mutation (ΔKPQ), which exhibits an enhanced persistent current⁽³⁾.
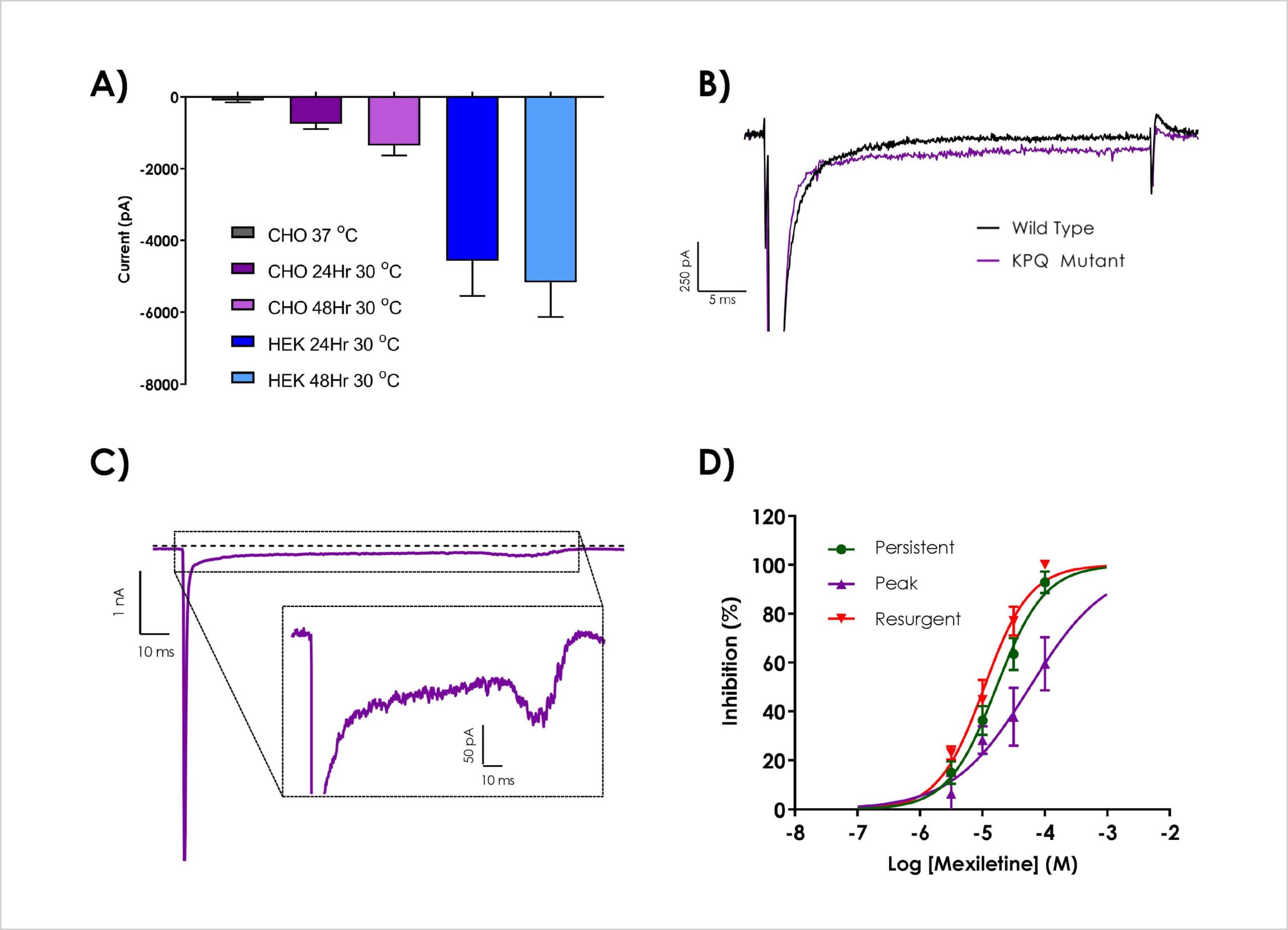
A) Polyclonal cell lines (CHO and HEK) were assessed for peak current size with varying days in culture at 30 °C. B) Side-by-side comparison of hNav1.5 WT and ΔKPQ cells using the QPatch 48 cell clone facility. C) Representative current traces elicited with the optimised voltage protocol, which show an increasing in current amplitude (cf. panel B). Pharmacological validation was performed, D) IC50 curves for mexiletine
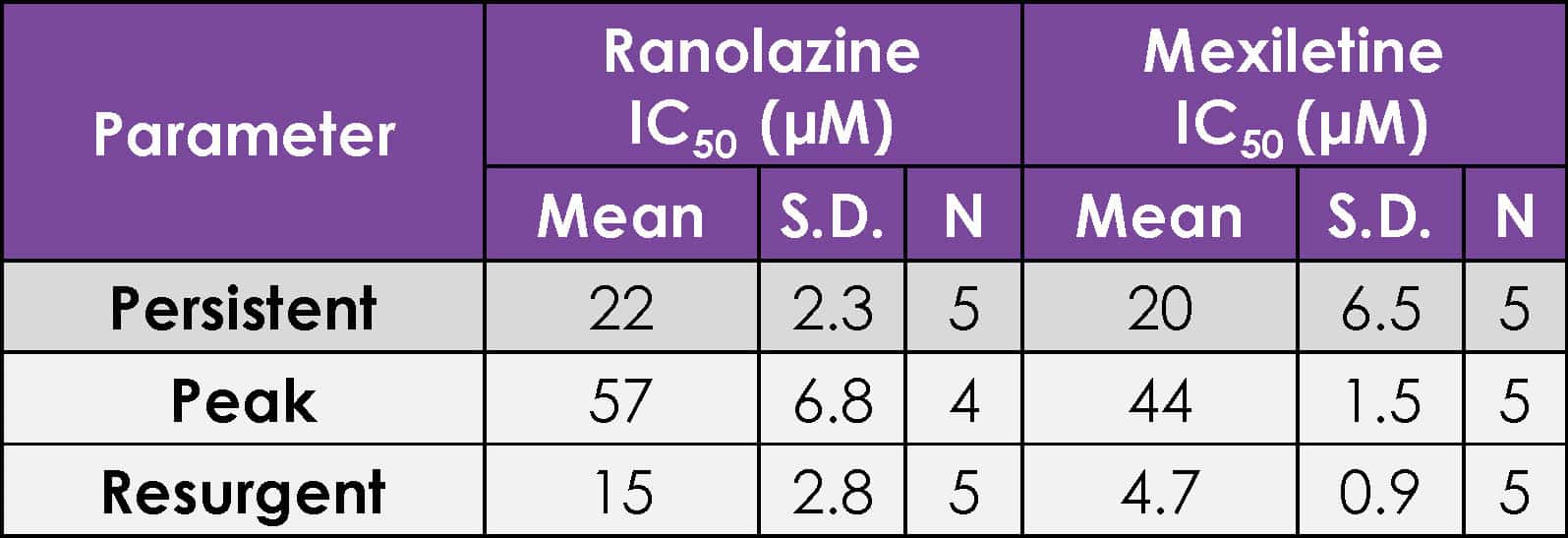
IC50 parameters for ranolazine and mexiletine against hNav1.5 ΔKPQ.
Dynamic hERG assay
Recent work by FDA and CiPA working groups indicate that addition of hERG kinetic data – obtained with the ‘Milnes’ voltage protocol (1) – to a modified ‘dynamic’ O’Hara-Rudy in silico model, improves proarrhythmic liability prediction (2). The kinetics of drug binding and unbinding to the hERG channel underlies compound potency, but there is evidence that compounds which become trapped in the pore of the channel carry a greater clinical risk (1). Up to now only high-fidelity manual patch clamp recordings have been used to reliably measure hERG channel binding kinetics and drug trapping, both important aspects of drug action and potency, as well as proarrhythmic liability.
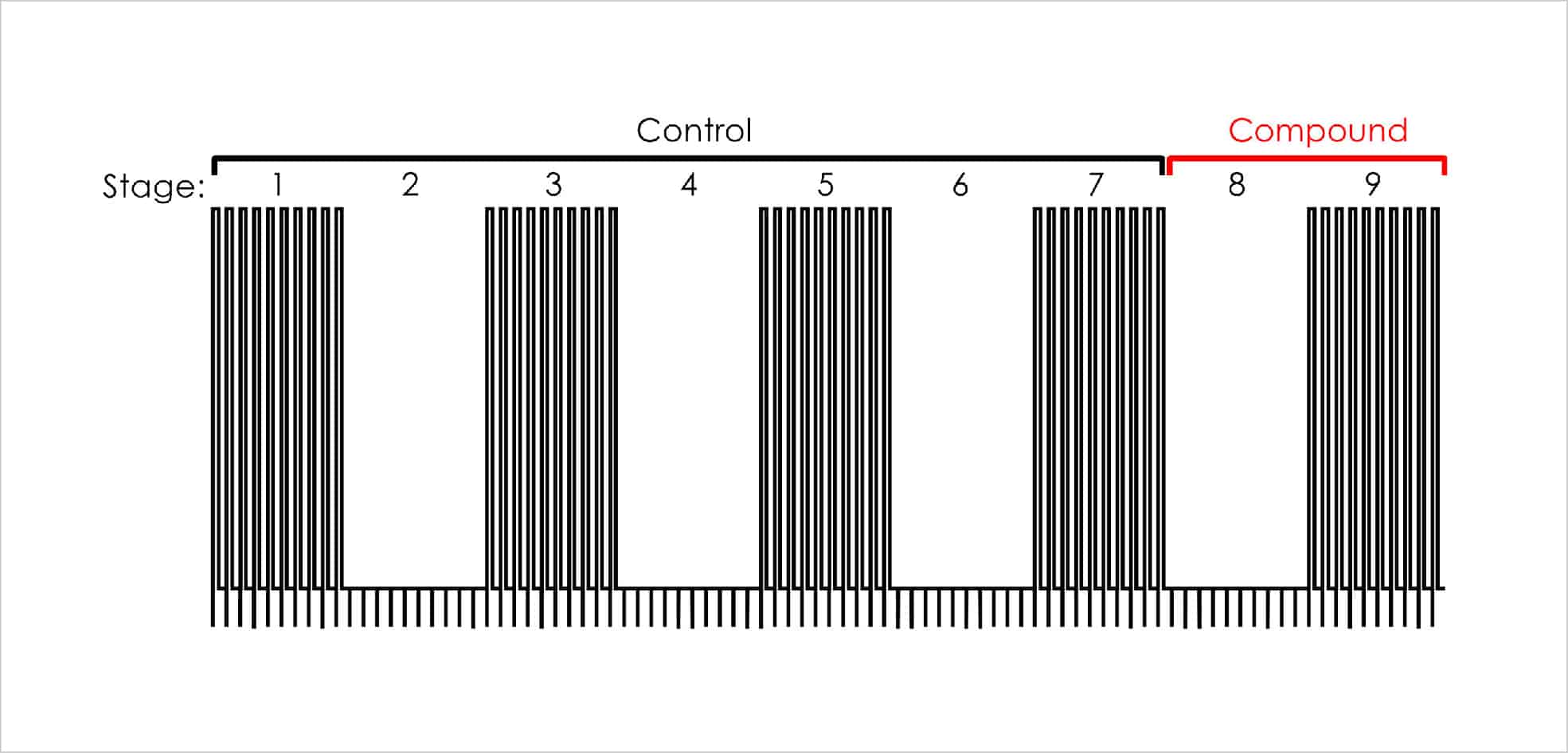
The Milnes protocol is comprised of 9 stages, each being 250 seconds long. Depolarising stages (1,3,5,7,9) consist of ten, 10 second depolarising steps with a sweep-to-sweep internal of 25 seconds.
Stable hERG currents in dynamic hERG QPatch assay
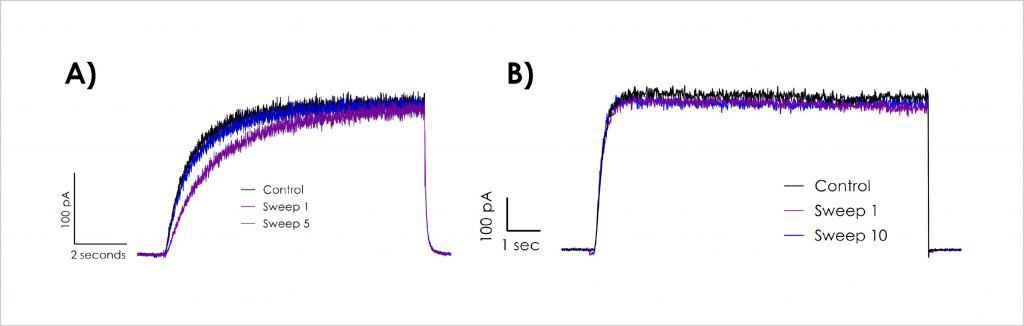
A) Initial experiments showed altered first pulse kinetics in stages (1,3,5,7,9) and significantly slower activation of hERG at room temperature compared to that at more physiological temperatures used in both Milnes et al.⁽¹⁾ and Li et al.⁽²⁾ B) Metrion’s optimised dynamic hERG assay with a stable current profile suitable for compound testing.
Dynamic hERG assay detects drug trapping
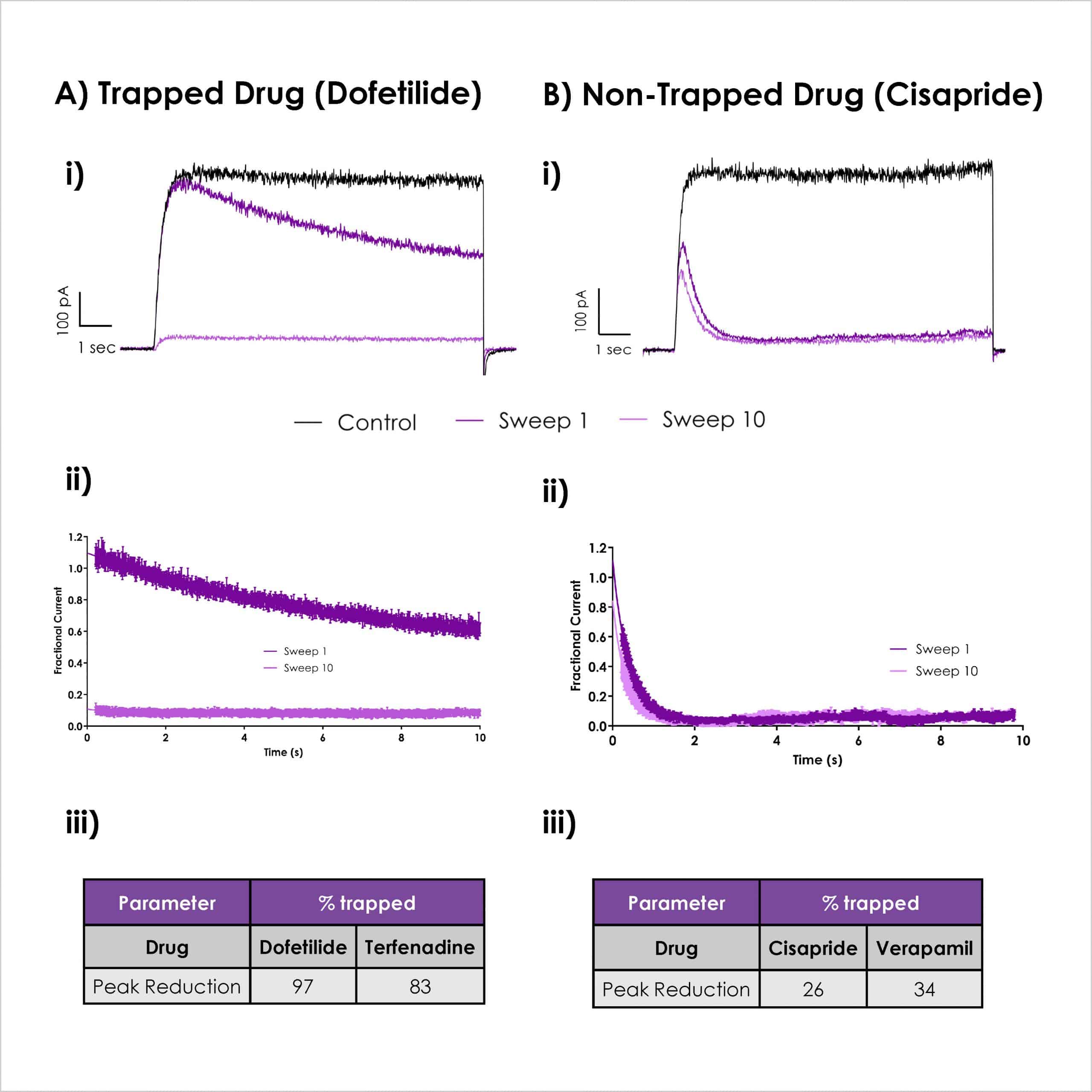
Ai) Representative traces for dofetilide (100 nM), ii) Average fractional hERG current for sweep 1 and 10 in the presence of dofetilide (100 nM), iii) Summary data of trapped drugs.
Bi) Representative traces for cisapride (300 nM), ii) Average fractional hERG current for sweep 1 and 10 in the presence of cisapride (300 nM). iii) Summary data table of non-trapped drugs.
Conclusions
Metrion have produced two additional QPatch 48 assays to improve its CiPA cardiac safety assay panel:
- hNav1.5 ΔKPQ ‘late’ current assay to reliably measure low amplitude persistent inward currents.
- Dynamic hERG assay to allow assessment of drug binding and trapping.
References
- Milnes et al. (2010). JPET. PMID: 20172036
- Li et al. (2017). Circ Arrhythm Electrophysiol. PMID: 28202629
- Chandra, Starmer and Grant (1998). Am J Physiol. PMID: 9612375
- Catterall et al. (2007). Toxicon. PMID: 17239913

Let’s work together
What are your specific ion channel and assay needs?
If you have any questions or would like to discuss your specific assay requirements, we will put you directly in touch with a member of our scientific team. Contact us today to discover more.
